

Welcome to the help files of
Part A - Sequence and Choices

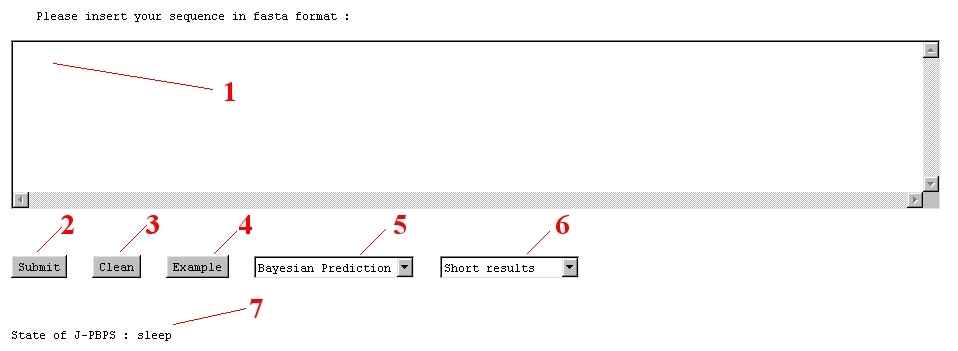
NB: Fasta format is a simple format : (1) the first line is the name of the protein beginning with a symbol ">" given ">name". (2) The following lines are used for the amino acid sequence. You could use the example button to see the example of protein 2aak.
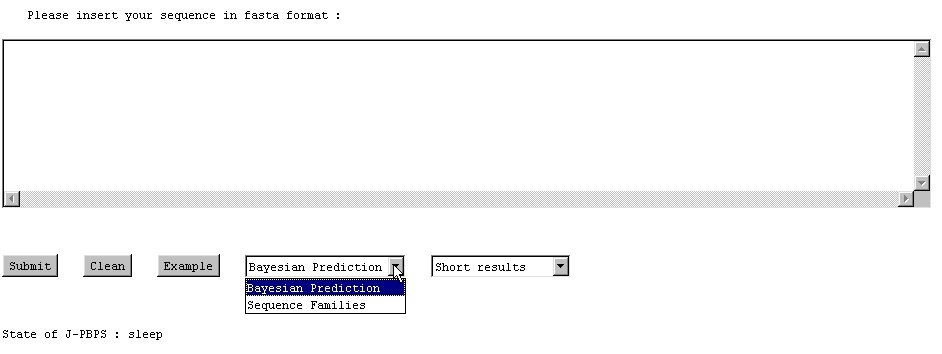
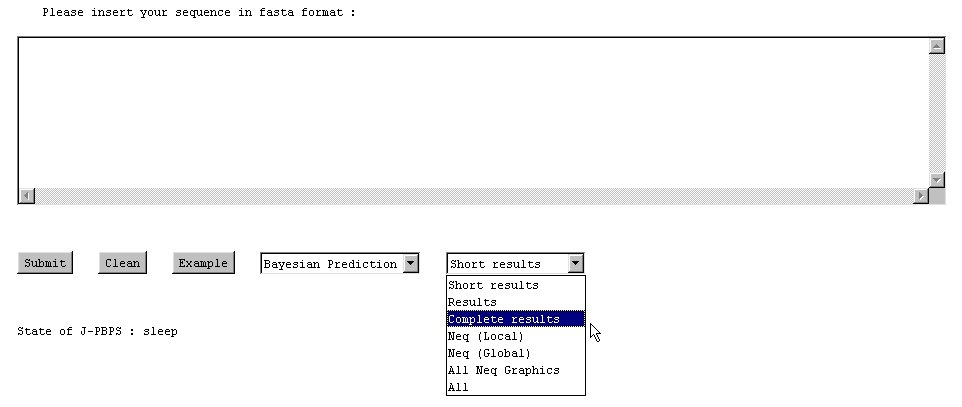
NB : The computation is always done in the same order (1) the results, (2) the local Neq and (3) the global Neq.