

Welcome to the help files of
Homology with Neq data

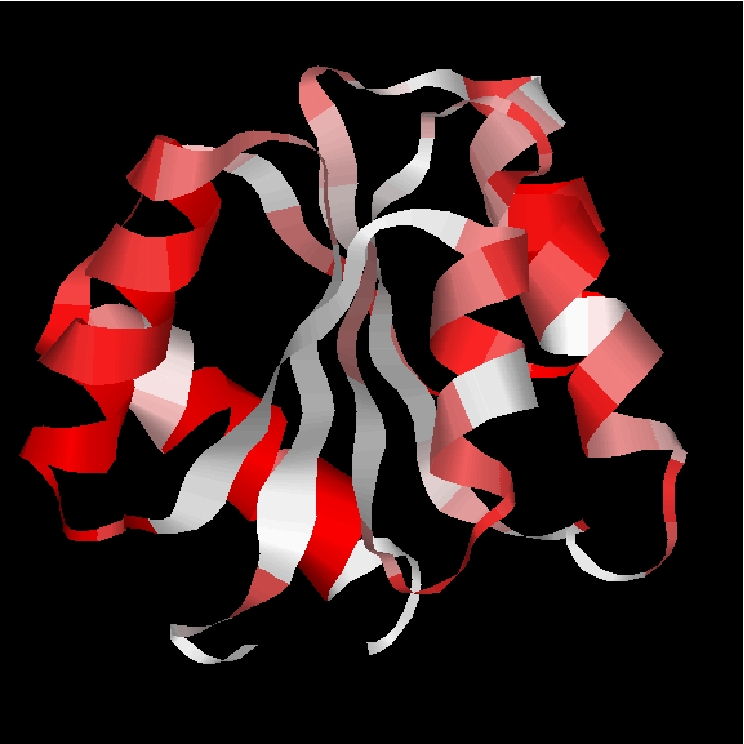
>1JBBA MASLPKRIIKETEKLVSDPVPGITAEPHDDNLRYFQVTIEGPEQSPYEDGI FELELYLPDDYPMEAPKVRFLTKIYHPNIDRLGRICLDVLKTNWSPALQIR TVLLSIQALLASPNPNDPLANDVAEDWIKNEQGAKAKAREWTKLYAKKKPE ________mdffppccbfbghiafdlffklpacdcddfebjkhpfklgopa cmmmdpgfipcfkbgciacdddddbehfklnopacfdldfnhfpfklopmm mmmmmklmmdpfbgkbccggklkfkmmmmmnopanmmmmmmmm________ >unknwon-Neq ________4566644534100012233443334533222214542243321 444453234223344465445446432343212234446555443234455 3333323455434122333665554434442122543333312________Simple prediction of 1JBBA.
1JBBA -MASLPKRIIKETEKLVSDPVPGITAEPHDDNLRYFQVTIEGPEQSPYEDGIFELELYLP
2AAK MSTPARKRLMRDFKRLQQDPPAGISGAPQDNNIMLWNAVIFGPDDTPWDGGTFKLSLQFS
:. **:::: ::* .** .**:. *:*:*: ::..* **:::*::.* *:*.* :.
2AAK-PB ZZfklmmmmmmmmmmmmmgcehiacdddfklckbccddddddehifkgoiacdddddddf
1JBBA-pred ________mdffppccbfbghiafdlffklpacdcddfebjkhpfklgopacmmmdpgf
1JBBA-pred ________456664453410001223344333453322221454224332144445323
1JBBA DDYPMEAPKVRFLTKIYHPNIDRLGRICLDVLKTNWSPALQIRTVLLSIQALLASPNPND
2AAK EDYPNKPPTVRFVSRMFHPNIYADGSICLDILQNQWSPIYDVAAILTSIQSLLCDPNPNS
:*** :.*.***:::::**** * ****:*:.:*** :: ::* ***:**..****.
2AAK-PB kbckbccddddfbdcddfklcfkopacdfklmmmpcfklcfklmmmmmmmmmmopcfklp
1JBBA-pred ipcfkbgciacdddddbehfklnopacfdldfnhfpfklopmmmmmmmklmmdpfbgkbc
1JBBA-pred 422334446544544643234321223444655544323445533333234554341223
1JBBA PLANDVAEDWIKNEQGAKAKAREWTKLYAKKKPE
2AAK PANSEAARMYSESKREYNRRVRDVVEQSWT----
* .:.*. : :.:: : :.*: .: .
2AAK-PB fblmlmmmmmmmmmlmmmmmmmmmmmmmZZ
1JBBA-pred cggklkfkmmmmmnopanmmmmmmmm________
1JBBA-pred 33665554434442122543333312________
Simple prediction of 1JBBA aligned with 2aak sequence (CLUSTALW), the traduction of 2aak in terms of Protein Blocks,
and the prediction of 1JBBA.
DPVPGI DPPAGI ** .** gcehia fbghia 410001Exemple of an intersting part of 1JBBA: in a conserved sequence, the Neq is strong, so PB series hia could be well-predict, the N-ter is less defined, but succession ghia is well coherent and PBs e and g are found in closest states.
FQVTIEGPEQ WNAVIFGPDD ::..* **:: cddddddehi cddfebjkhp 3322221454An other intersting exemple: the succession seems to be really coming from a beta-sheet , i.e. PB's series d, but in 1JBBA, the repetitive structure seems to be shorter.
back
Last modif : 25 April 2004