

Prediction of local structures from the sequence
1- The sequence fragment is "GNPSTVQNPRLMNP".
2- The normalized occurence matrix associated with PB x is taken. It is defined by fij the frequence of amino acid i in position j divided by the frequence of amino acid i in the databank (i is in the range 1 to 20 and j in the range -7 to +7, the sequence window is always 15 residue length).
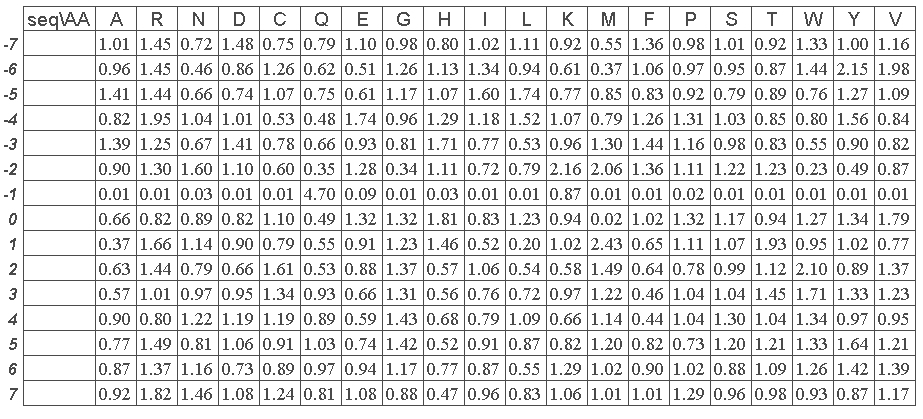
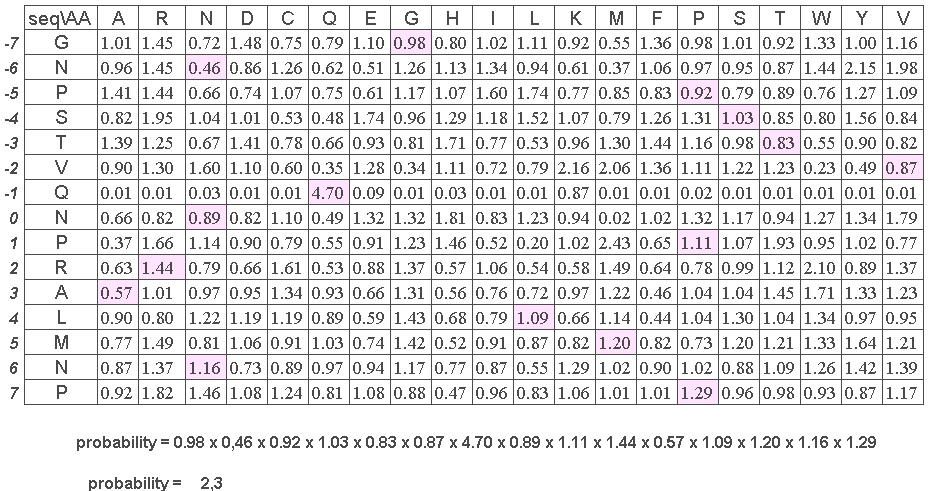
| Protein Blocks | Score | Normalized Scores | |
| a | 1.93 | 0.23 | |
| b | 0.12 | 0.01 | |
| c | 0.23 | 0.03 | |
| d | 0.02 | 0.00 | |
| e | 0.14 | 0.02 | |
| f | 0.25 | 0.03 | |
| g | 0.56 | 0.07 | |
| h | 0.56 | 0.07 | |
| i | 0.89 | 0.11 | |
| j | 0.63 | 0.08 | |
| k | 0.45 | 0.05 | |
| l | 0.12 | 0.01 | |
| m | 0.05 | 0.01 | |
| n | 0.08 | 0.01 | |
| o | 0.97 | 0.12 | |
| p | 1.23 | 0.15 | |
| sum = | 8.23 | 1.00 |
5- The 16 scores are re-arranged :
a, p, o, i, j, g, h, k, f, c, e, b, l, n, m, d.
Here the most probable PB is the PB a.
6- If the Neq is small (close to 1), the PB a has a good probabilty to be the real good. On the contrary, if the Neq
is high, the sequence use are not informative, so it is more interessting to select not only the most probable PB, but a set of probable PBs like
PBs a, p, o, i and j. The Neq value and the desired prediction rate would tell you the exact number of PB to be select.
7- The prediction rate is computed as the sum of well predicted BPs (true PBs found the most probable / total number of PBs).
back
Last modif : 25 April 2004