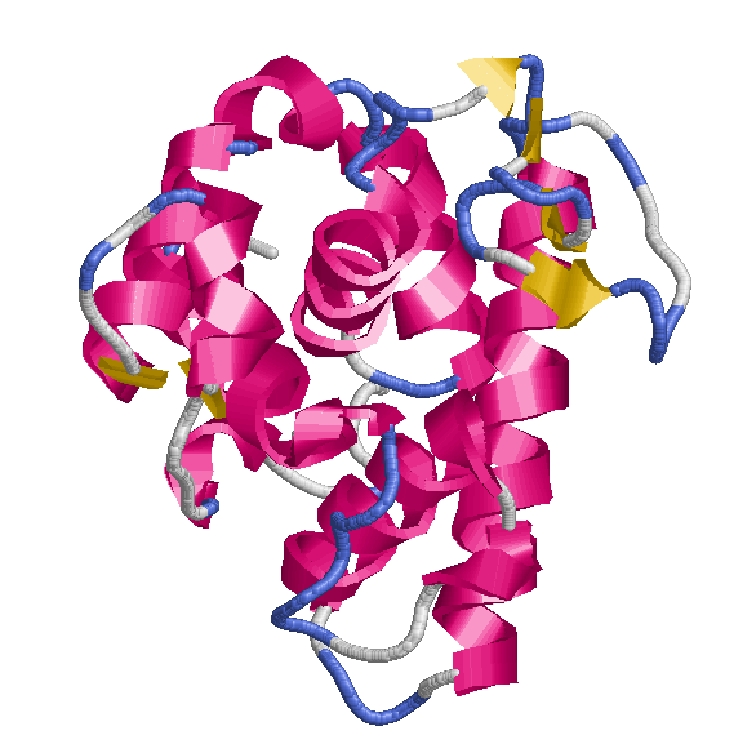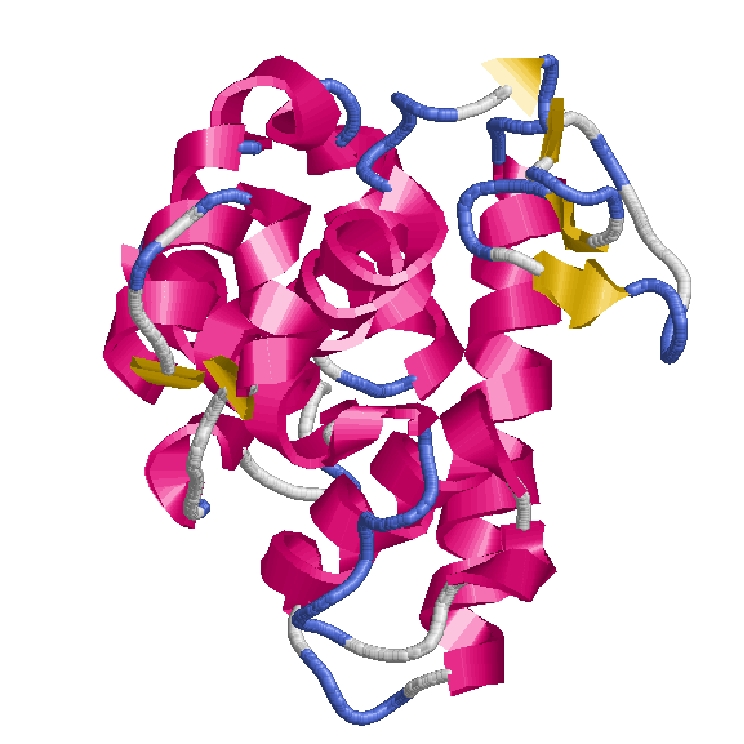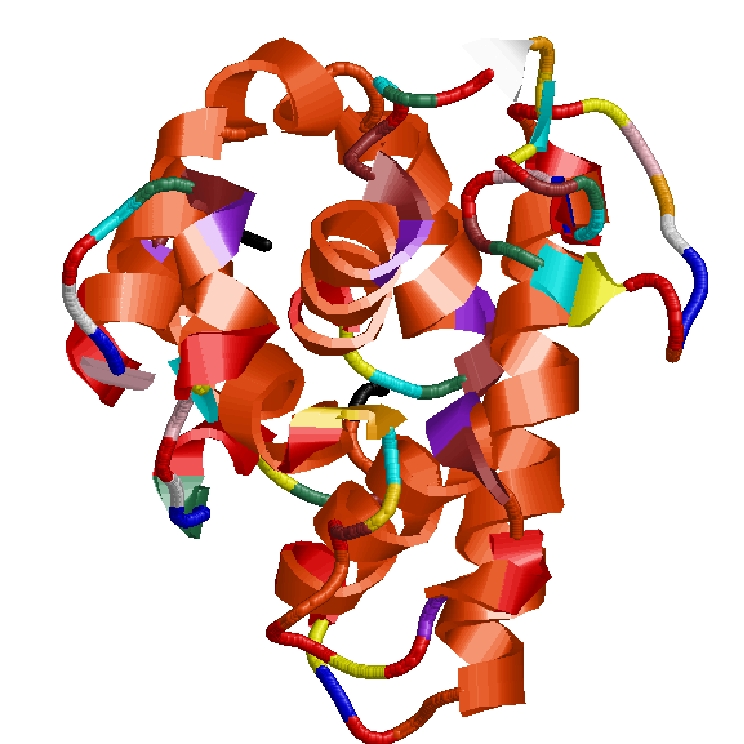

How is a 3D protein structure coded into Protein Blocks ?
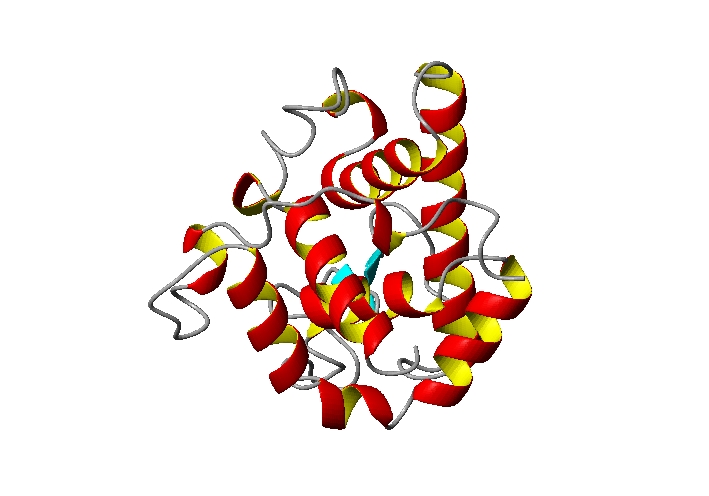

HEADER HYDROLASE(O-GLYCOSYL) 05-MAY-94 153L 153L 2 ATOM 1 N ARG 1 6.350 34.124 50.750 1.00 41.90 153L 116 ATOM 2 CA ARG 1 6.324 32.707 50.379 1.00 54.68 153L 117 ATOM 3 C ARG 1 6.334 32.484 48.874 1.00 14.63 153L 118 ATOM 4 O ARG 1 7.356 32.060 48.316 1.00 23.75 153L 119 ATOM 5 CB ARG 1 5.009 32.300 50.934 1.00 40.20 153L 120 ATOM 6 CG ARG 1 4.526 33.584 51.604 1.00 56.81 153L 121 ATOM 7 CD ARG 1 3.012 33.793 51.724 1.00 64.62 153L 122 ATOM 8 NE ARG 1 2.515 34.238 50.431 1.00 63.14 153L 123 ATOM 9 CZ ARG 1 2.352 33.394 49.423 1.00 37.38 153L 124 ATOM 10 NH1 ARG 1 2.588 32.086 49.557 1.00 56.33 153L 125 ATOM 11 NH2 ARG 1 1.895 33.858 48.262 1.00 59.78 153L 126 ATOM 12 N THR 2 5.206 32.767 48.261 1.00 15.22 153L 127 ATOM 13 CA THR 2 5.197 32.618 46.826 1.00 15.40 153L 128 ATOM 14 C THR 2 4.781 33.870 46.108 1.00 23.28 153L 129 ATOM 15 O THR 2 4.716 33.930 44.845 1.00 20.96 153L 130 ATOM 16 CB THR 2 4.452 31.426 46.229 1.00 12.40 153L 131 ATOM 17 OG1 THR 2 3.089 31.502 46.538 1.00 20.80 153L 132 ATOM 18 CG2 THR 2 5.066 30.133 46.701 1.00 26.07 153L 133 ATOM 19 N ASP 3 4.497 34.900 46.908 1.00 31.19 153L 134 ATOM 20 CA ASP 3 4.020 36.132 46.259 1.00 35.11 153L 135 ATOM 21 C ASP 3 4.987 37.273 45.992 1.00 19.94 153L 136 ATOM 22 O ASP 3 4.530 38.398 45.795 1.00 29.83 153L 137 ATOM 23 CB ASP 3 2.951 36.717 47.185 1.00 28.17 153L 138 ATOM 24 CG ASP 3 3.632 36.891 48.524 1.00 26.63 153L 139 ATOM 25 OD1 ASP 3 4.821 36.990 48.715 1.00 43.06 153L 140 ATOM 26 OD2 ASP 3 2.844 36.730 49.510 1.00 54.69 153L 141 ATOM 27 N CYS 4 6.285 37.076 46.089 1.00 17.27 153L 142 ATOM 28 CA CYS 4 7.131 38.210 45.919 1.00 16.83 153L 143 ATOM 29 C CYS 4 7.073 38.939 44.605 1.00 15.34 153L 144 ATOM 30 O CYS 4 7.459 40.095 44.530 1.00 22.22 153L 145 ATOM 31 CB CYS 4 8.597 37.815 46.045 1.00 14.54 153L 146 ATOM 32 SG CYS 4 8.982 37.124 47.655 1.00 23.95 153L 147 ATOM 33 N TYR 5 6.691 38.264 43.538 1.00 11.91 153L 148 ATOM 34 CA TYR 5 6.719 38.935 42.270 1.00 13.01 153L 149 ATOM 35 C TYR 5 5.366 39.179 41.653 1.00 13.67 153L 150 ATOM 36 O TYR 5 5.254 39.476 40.486 1.00 19.57 153L 151 ATOM 37 CB TYR 5 7.599 38.124 41.282 1.00 21.26 153L 152 ATOM 38 CG TYR 5 9.055 37.997 41.772 1.00 17.97 153L 153 ATOM 39 CD1 TYR 5 9.974 39.045 41.627 1.00 28.24 153L 154 ATOM 40 CD2 TYR 5 9.485 36.843 42.432 1.00 16.83 153L 155 ATOM 41 CE1 TYR 5 11.281 38.964 42.115 1.00 29.20 153L 156 ATOM 42 CE2 TYR 5 10.780 36.732 42.944 1.00 19.39 153L 157 ATOM 43 CZ TYR 5 11.672 37.794 42.763 1.00 27.35 153L 158 ATOM 44 OH TYR 5 12.956 37.704 43.225 1.00 26.92 153L 159 ATOM 45 N GLY 6 4.320 39.020 42.427 1.00 19.79 153L 160 ATOM 46 CA GLY 6 2.986 39.221 41.892 1.00 15.13 153L 161 ATOM 47 C GLY 6 2.071 38.002 41.992 1.00 17.68 153L 162 ATOM 48 O GLY 6 2.522 36.960 42.438 1.00 21.96 153L 163 ATOM 49 N ASN 7 0.819 38.158 41.541 1.00 18.97 153L 164 ATOM 50 CA ASN 7 -0.269 37.184 41.565 1.00 20.89 153L 165 ATOM 51 C ASN 7 -0.922 37.069 40.232 1.00 13.79 153L 166 ATOM 52 O ASN 7 -1.584 37.990 39.760 1.00 14.39 153L 167 ATOM 53 CB ASN 7 -1.372 37.625 42.536 1.00 19.21 153L 168 ATOM 54 CG ASN 7 -2.418 36.535 42.784 1.00 42.06 153L 169 ATOM 55 OD1 ASN 7 -2.908 35.750 41.931 1.00 29.02 153L 170 ATOM 56 ND2 ASN 7 -2.761 36.491 44.049 1.00 54.80 153L 171 END 153L1736
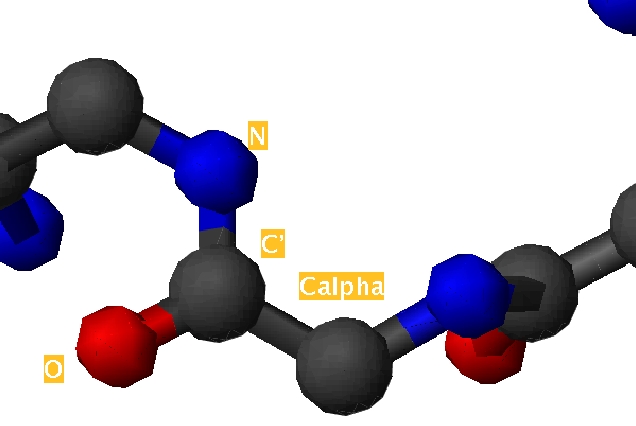
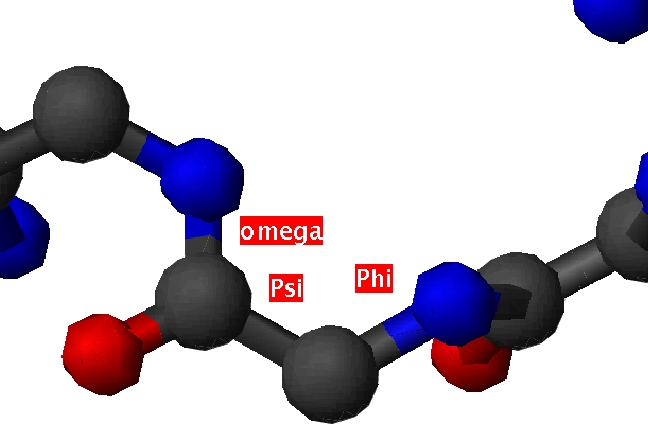
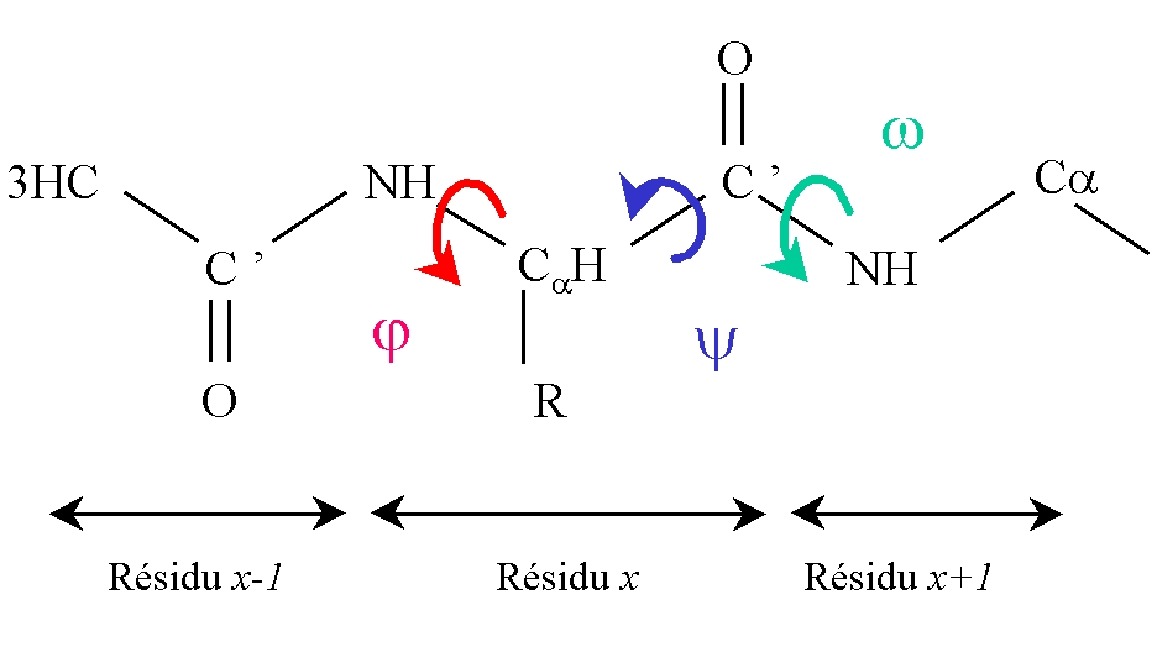
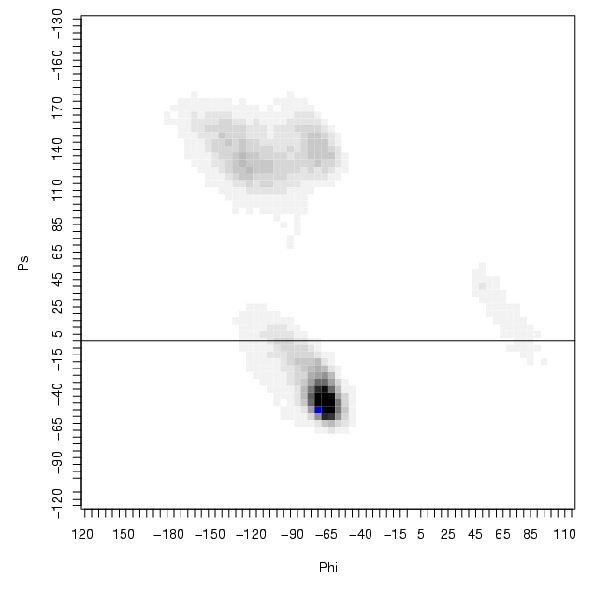
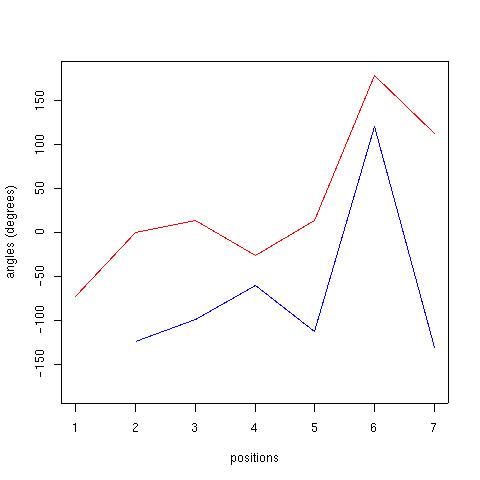
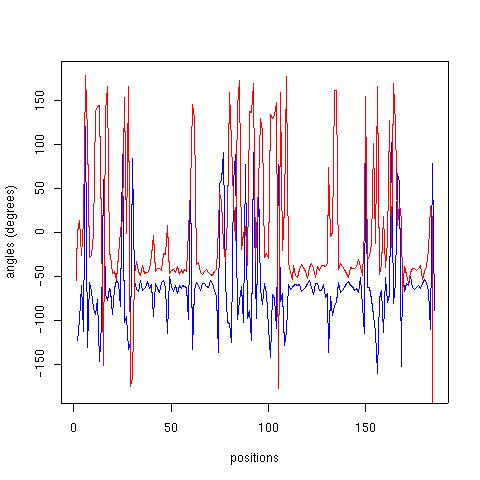
| Position | Phi | Psi |
| 1 | --- | -73.17 |
| 2 | -124.40 | 0.21 |
| 3 | -98.44 | 13.49 |
| 4 | -60.37 | -25.89 |
| 5 | -112.84 | 13.68 |
| 6 | 120.99 | 178.29 |
| 7 | -130.63 | 112.67 |
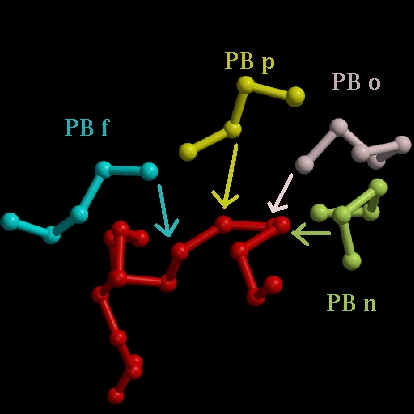
1 (A) RTDCYGNVNRIDTTGASCKTAKPEGLSYCGVSASKKIAE (B) ZZmnopfklpccebjafklmmmnopabecjklmmmmmmm (C) cccccccccccccccccaaaaaaaccccccaaaaaaaaa (D) CCTTTTCGGGCCCCCBCHHHHGGGCCCCCBHHHHHHHHH 41 (A) DLQAMDRYKTIIKKVGEKLCVEPAVIAGIISRESHAGKV (B) mmmmmmmmmmmmmmmmmnopafklmmmmmmmmnooolap (C) acaaaaaaaaaaaaaaaaacccaaaaaaaaaaacccccc (D) HHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHGGGTT 81 (A) KNGWGDRGNGFGLMQVDKRSHKPQGTWNGEVHITQGTTI (B) ehiafkopagcjkopafklmccehjfklmklmmmmmmmm (C) cccccccccccccccccccccccccccccaaaaaaaaaa (D) BTTBTTTTCEETTTTEETTTTCCCCTTTTHHHHHHHHHH 121 (A) INFIKTIQKKFPSWTKDQQLKGGISAYNAGAGNVRSYAR (B) mmmmmmmmmmmmbcfklmmmmmmmmmmnomklmnbfklm (C) aaaaaaaaaacccccaaaaaaaaaaaaaccccccccccc (D) HHHHHHHHHHTTTTCHHHHHHHHHHHHHHCGGGCCTTTT 161 (A) DIGTTHDDYANDVVARAQYYKQHGY (B) goiahilmmmmmmmmmmmmmmnoZZ (C) ccccccccaaaaaaaaaaaaaaacc (D) TTTTTTTTHHHHHHHHHHHHHHCCC